Projects
The Harvard Medical School-based Center for Cancer Systems Pharmacology is an NCI Cancer Systems Biology Center of Excellence that studies responsiveness and resistance to anti-cancer drugs. The Center focuses on targeted small molecule therapies and newly emerging immune checkpoint inhibitors (ICIs), two cornerstones of precision cancer medicine. Center members are trying to identify which patients will benefit from each class of drugs and why cancers that are initially drug-sensitive often become resistant. The Center also studies drug toxicity, with the aim of reducing the burden of therapy for individual patients.
Our research is performed using a mix of experimental and computational approaches and follows a bedside to bench and bench to bedside paradigm. We are committed to the generation of reproducible and publicly accessible data and to open-source software.
The Center for Cancer Systems Pharmacology involves three inter-linked research projects, a systems pharmacology core, and multiple education and outreach activities.
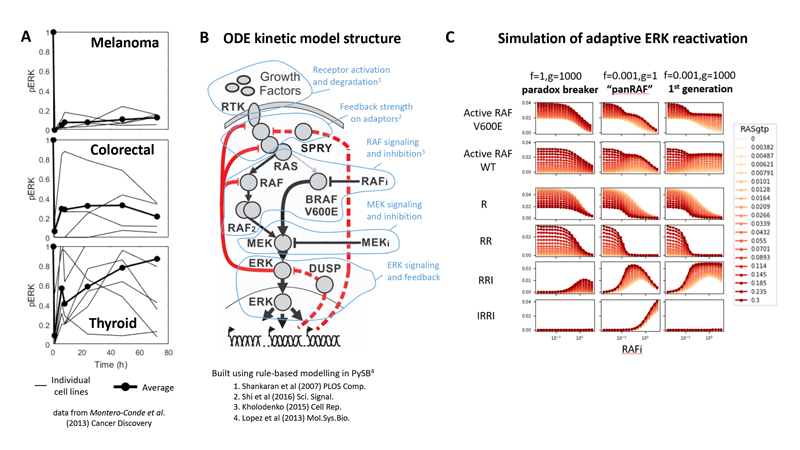
B) Structure of the kinetic model explaining adaptive ERK reactivation in BRAF-mutant cancers.
C) Simulations predicting the efficacy of different classes of RAF inhibitors (1st generation, paradox breaker and pan-RAF inhibitors) on adaptive ERK reactivation.